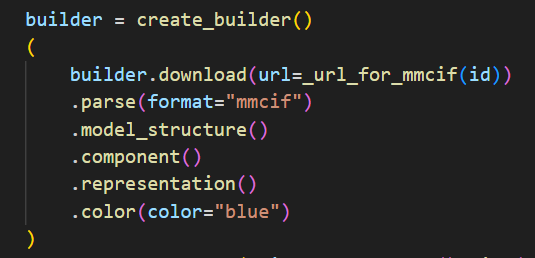
Mol* (/'molstar/) is a modern web-based open-source toolkit for visualisation and analysis of large-scale molecular data
Interactive Examples
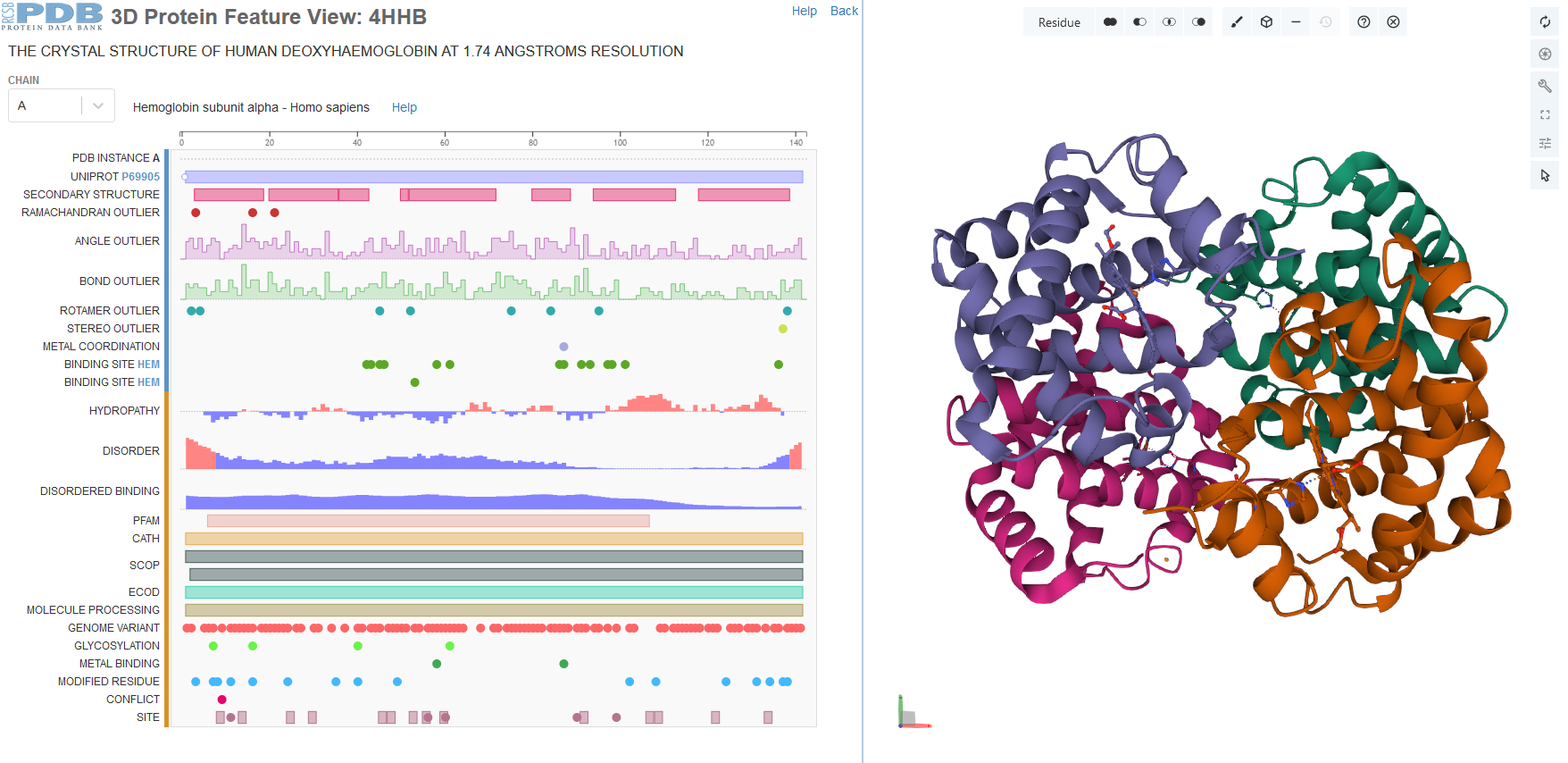
CellPack model of enveloped HIV capsid.
Opens in the Mesoscale Explorer
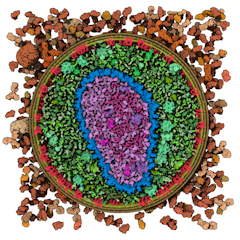
GAIN domain tethered agonist exposure (doi:10.1016/j.molcel.2020.12.042). Based on an animation by Ramon Guixà-González.
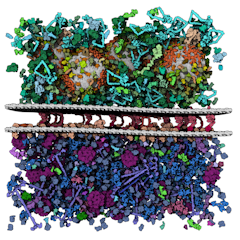
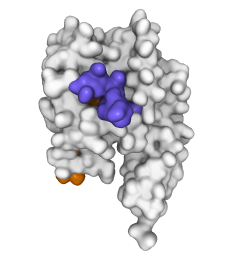
Petworld model of the synaptic gap.
Opens in the Mesoscale Explorer
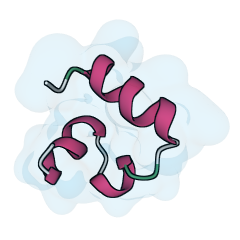
Villin folding trajectory by Stefan Doerr (doi:10.6084/m9.figshare.12040257.v1).
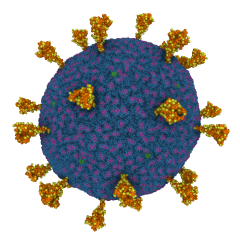
SARS-CoV-2 Virion. Coarse-grained model by Alvin Yu et al. (doi:10.1016/j.bpj.2020.10.048).

BtuB molecules in a lipid bilayer. Coarse-grained model by Matthieu Chavent et al. (doi:10.1038/s41467-018-05255-9).
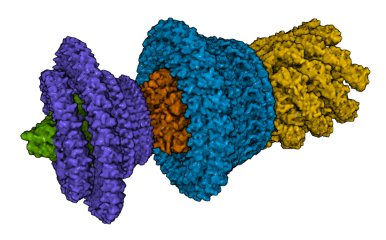
Cryo-EM structure of the bacterial flagellar motor-hook complex (@Jiaxing_Tan_ et al. PDB 7CGO, doi:10.1016/j.cell.2021.03.057).
David Sehnal, Sebastian Bittrich, Mandar Deshpande, Radka Svobodová, Karel Berka, Václav Bazgier, Sameer Velankar, Stephen K Burley, Jaroslav Koča, Alexander S Rose: Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures, Nucleic Acids Research, 2021; 10.1093/nar/gkab31.
Apps, Extensions & Features
Community Highlights